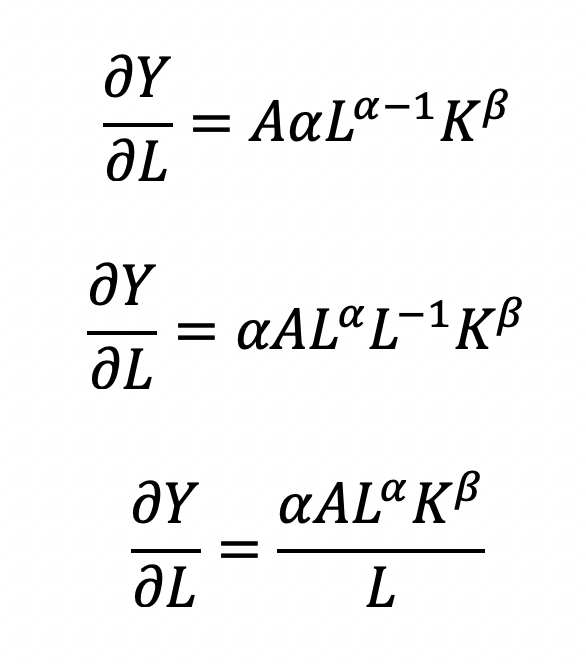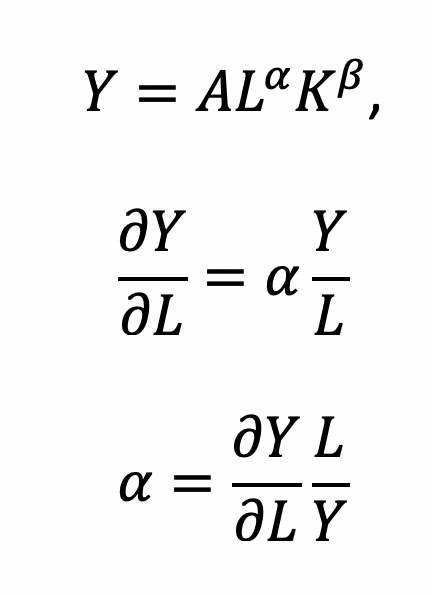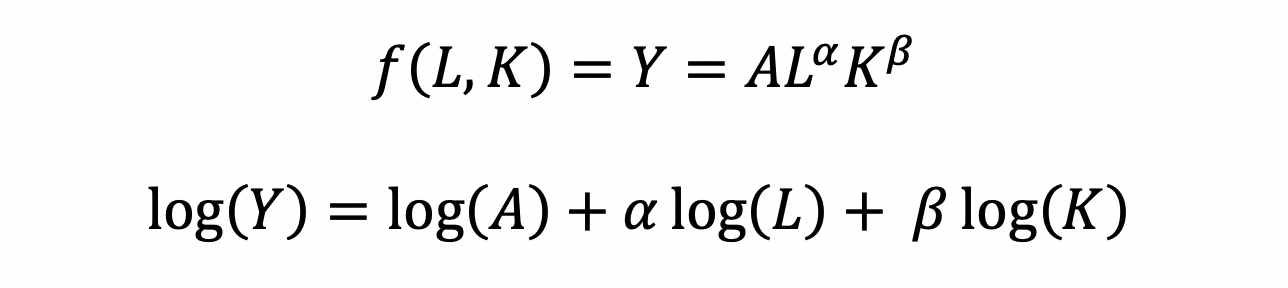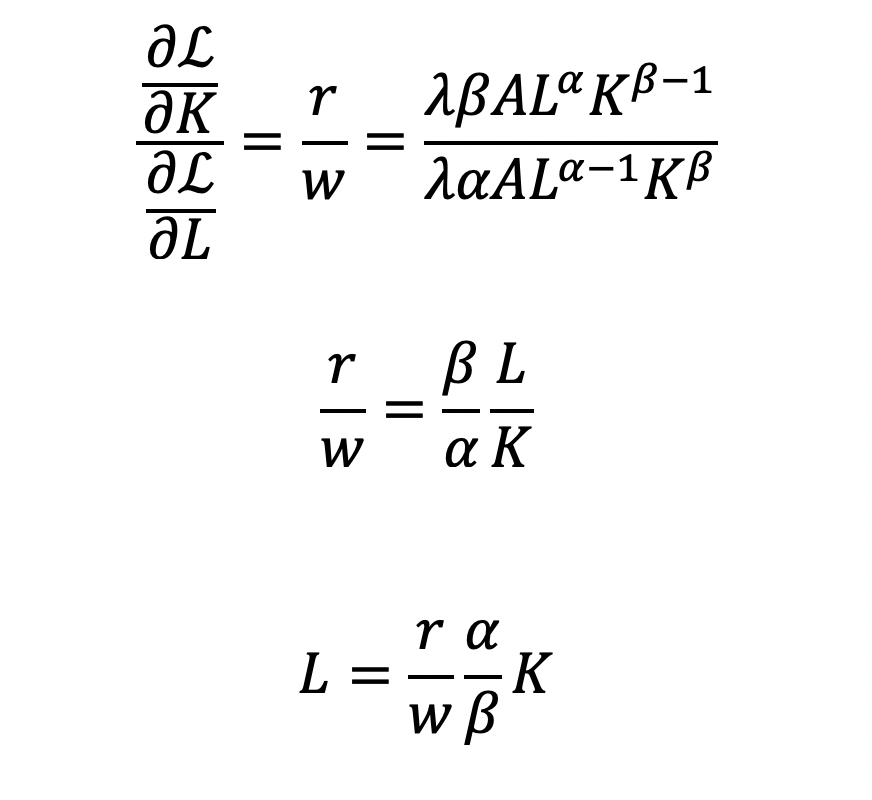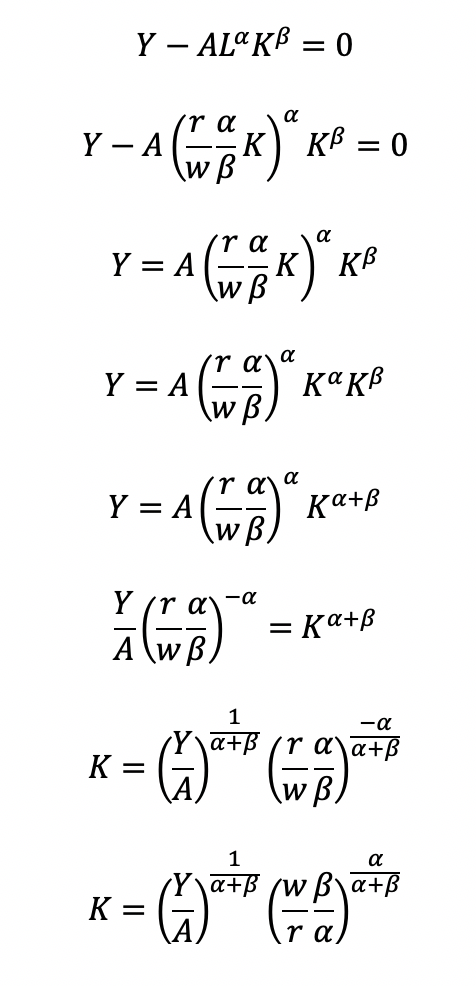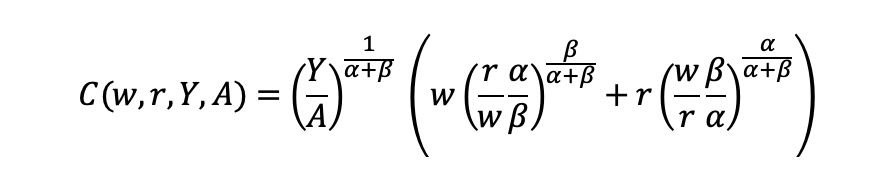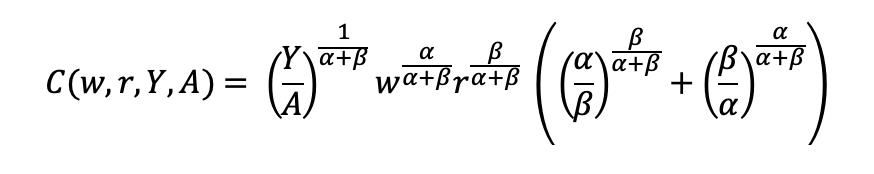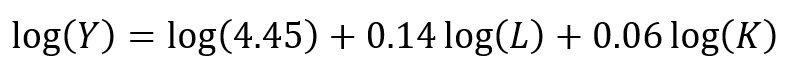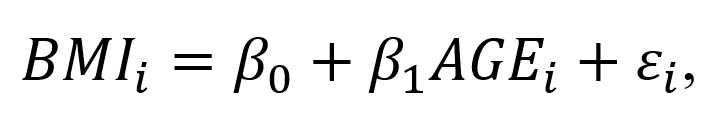Update 2: This article was updated on 12 August 2023 when Dimanjan Dahal (Twitter account) identified a better way to present the Lagrangian functions. I updated this to better reflect the minimization problem and set the partial derivative solution to 0. Thank you, Dimanhan.
Update 1: This article was updated on 11 October 2021 when an anonymous reader identified an error with the example used at the end. The error was the negative value generated for the output elasticity of capital. In the previous example, I used R to generate a set of random numbers that were used in a regression model. The beta coefficient generated a negative value which was used in the linear form of the Cobb-Douglass equation. Since the output of elasticity should be between the values of 0 and 1, this negative coefficient should not be possible. Hence, I’ve updated the data frame used in the example to avoid this issue. Appreciation goes out to the anonymous reader who identified this error.
INTRODUCTION
The Cobb-Douglas (CD) production function is an economic production function with two or more variables (inputs) that describes the output of a firm. Typical inputs include labor (L) and capital (K). It is similarly used to describe utility maximization through the following function [U(x)]. However, in this example, we will learn how to answer a minimization problem subject to (s.t.) the CD production function as a constraint.
The functional form of the CD production function:
where the output Y is a function of labor (L) and capital (K), A is the total factor productivity and is otherwise a constant, L denotes labor, K denotes capital, alpha represents the output elasticity of labor, beta represents the output elasticity of capital, and (alpha + beta = 1) represents the constant returns to scale (CRS). The partial derivative of the CD function with respect to (w.r.t) labor (L) is:
Recall that quantity produced is based on the labor and capital; therefore, we can solve for alpha:
This will yield the marginal product of labor (L). If alpha = 2, then a 10% increase in labor (L) will result in a 20% increase in output (Y).
The partial derivative of the CD function with respect to (w.r.t) labor (K) is:
This will yield the marginal product of capital (K).
The CD production function can be converted to a linear model by taking the logarithm of both sides of the equation:
This will allow for OLS regression methods, which is commonly used in economics to understand the association between inputs (L and K) on production (Y).
However, what happens when we are interested in the marginal cost with respect to (w.r.t.) production (Y)? This becomes a constraint (cost) minimization problem where the firm can control how much L and K they will use. In other words, we want to minimize the cost subject to (s.t.) the output
Cost becomes a function of wage (w), the amount of labor (L), price of capital (r), and the amount of capital (K). To determine the optimal amount of inputs (L and K), we solve this minimization constraint using the Lagrange multiplier method:
Solve for L
Substitute L in the constraint term (CD production function) in order to solve for K
Now, we can completely solve for L (as a function of Y, A, w, and r) by substituting for K
Substitute L and K into the cost minimization problem
Simplify
Final cost function
Let’s see how we can use the results from a regression model to give us information about the total costs w.r.t. to the quantity produced.
Recall the linear form of the Cobb-Douglas production function:
I simulated some data where we have the capital, labor, and quantity produced in R.
## Use the following libraries: library(jtools) library(broom) library(ggstance) library(broom.mixed) ## Generate random data for the data frame (cddata) set.seed(1234) production <- sample(100:600, 30, replace=TRUE) labor <- sample(50:350, 30, replace=TRUE) capital <- sample(6000:7000, 30, replace=TRUE) ## Cost function parameters: wage and price constants wage <- 35.00 price <- 30.00 ## Set up the data frame (cddata): cddata <- data.frame(production = production, labor = labor, capital = capital, wage = wage, price = price) ## Name rows using some timeline from 1988 to 2017 (30 years for 30 observations for each variable): row.names(cddata) <- 1988:2017
Then I perform a regression model using OLS
## Setting up the model, where log(a) is eliminated due to it being the intercept. cd.lm <- lm(formula = log(production) ~ log(labor) + log(capital), data = cddata) summary(cd.lm) Residuals: Min 1Q Median 3Q Max -0.96586 -0.25176 0.06148 0.37513 0.67433 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.44637 17.41733 0.255 0.800 log(labor) 0.14373 0.23595 0.609 0.548 log(capital) 0.05581 2.00672 0.028 0.978 Residual standard error: 0.5065 on 27 degrees of freedom Multiple R-squared: 0.01414, Adjusted R-squared: -0.05888 F-statistic: 0.1937 on 2 and 27 DF, p-value: 0.8251
After running the model, I stored the coefficients for use later in the production function.
## Store the coefficients coeff <- coef(cd.lm) ## Assign the values to the production function parameters where Y = AL^(alpha)K^(beta) intercept <- coeff[1] alpha <- coeff[2] beta <- coeff[3]
From the parameters, we can get A (intercept), alpha (log(labor)), and beta (log(capital)).
This will give us the quantity produced (Y) for given data on labor (L) and capital (K).
We can get the total costs (C) based on the quantity produced (Y) using the cost function:
I set up my R code so that I have the intercept, alpha, beta, labor, wage, and price of the capital set up. I estimated each part of the cost function separately and then multiply the parts at the end.
## Cost PartA <- (production / intercept)^(1 / alpha + beta) PartB <- wage^(alpha / alpha + beta) PartC <- price^(beta / alpha + beta) PartD <- as.complex(alpha / beta )^(beta / alpha + beta) + as.complex(beta/ alpha)^(alpha / alpha + beta) costs <- PartA * PartB * PartC * PartD
Note: R has a problem with performing complex operations with exponents that were defined using arrays or vectors. If you try to compute something like x^{alpha}, you will get an error where the value is “NaN.” I don’t have a complete understanding of the problem, but the solution is to make sure your root or base term is preceded by “as.complex(x)” to resolve the issue.I plot the relationship between quantity produced and cost. In other words, this tells us the lowest costs needed to produce the quantities on the plot.
plot(production, costs)
CONCLUSIONS
Using the Cobb-Douglas production function and the cost minimization approach, we were able to find the optimal conditions for the cost function and plot the outcome relative to the quantity produced. As production increases, the minimum cost needed increases in a non-linear, exponential fashion, which makes sense given that Y (quantity produced) is in the numerator on the right-hand side of the cost function and positively related to the cost.
This was a fun exercise that made me think about the usefulness of the Cobb-Douglas production function, which I learned to optimize multiple times in my Economics courses. I was excited to find a pleasant utility for it using simulated data and will probably explore more exercises like this in the future.
REFERENCEs
I used a lot of resources to write this blog, which are provided below.
A site dedicated to the discussion of economics called EconomicsDiscussion.net was a great resource.
These papers were incredibly helpful in preparing the example in R:
Lin CP. The application of Cobb-Douglas production cost functions to construction firms in Japan and Taiwan. Review of Pacific Basin Financial Markets and Policies Vol. 5, No. 1 (2002): 111–128.
Larriviere JB, Sandler R. A student friendly illustration and project: empirical testing of the Cobb-Douglas production function using major league baseball. Journal of Economics and Economic Education Research, Volume 13, Number 3, 2012: 81-92
Hu, ZH. Reliable Optimal Production Control with Cobb-Douglas Model. Reliable Computing. 1998; 4(1): 63-69.
I encountered some issues regarding complex numbers in R. Fortunately, I found some great resources about it.
I found a great discussion about R’s calculation of exponents and “NaN” results and why complex numbers can mess up your math in R.
Another good site (R Tutorial: An Introduction to Statistics) explaining complex numbers in R.
John Myles White wrote a nice article about complex numbers in R.
Acknowledgements: I would like to thank the user who reached out to me about the coefficient errors for the output elasticity of capital. This helps me to learn my mistakes and correct them. Without the support and guidance from the community, I would not achieve my own goals of being a lifelong learner. Thank you.