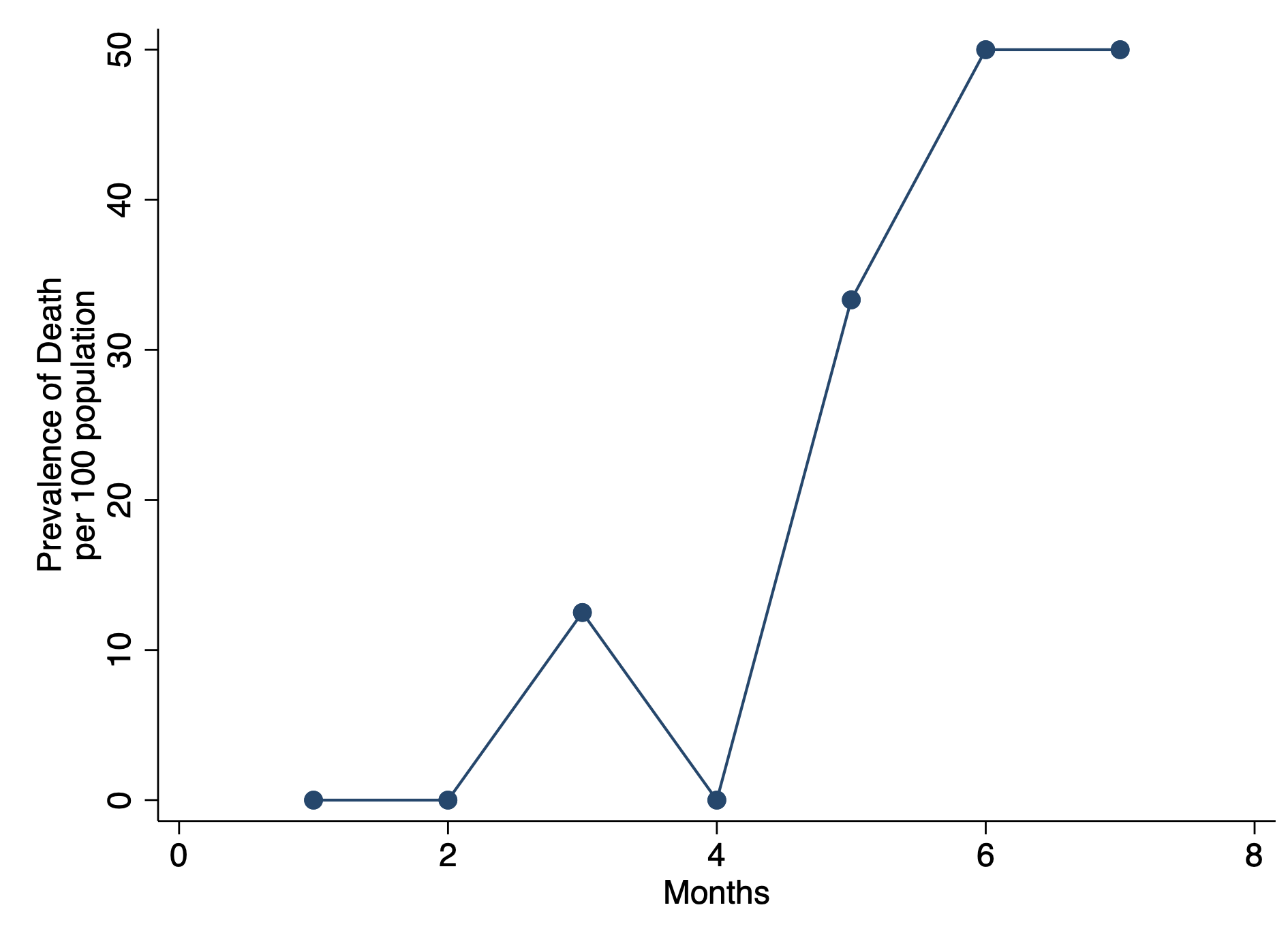
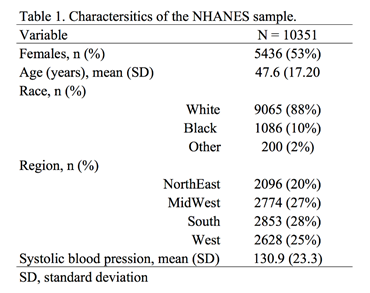
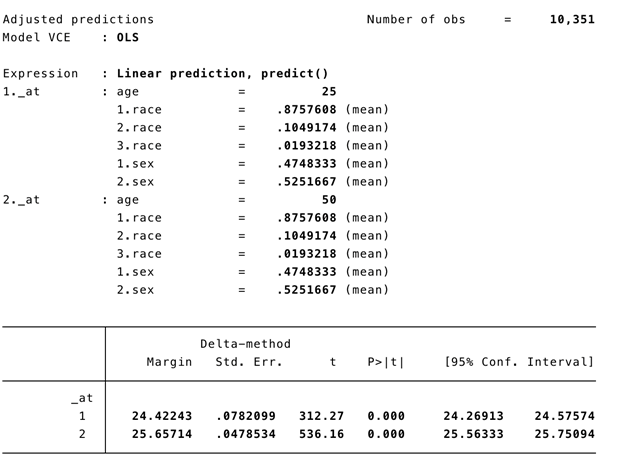
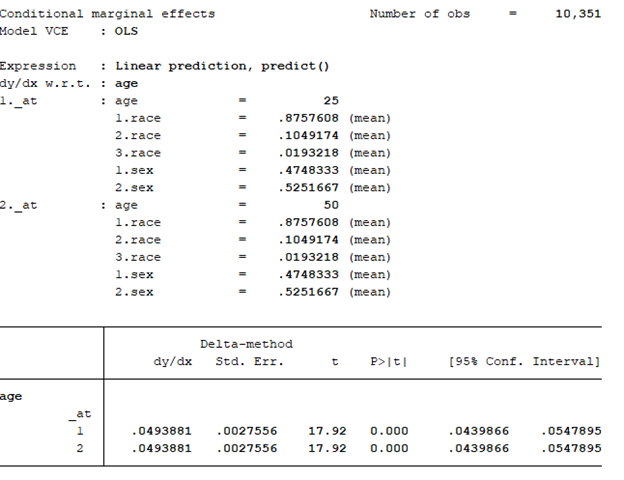
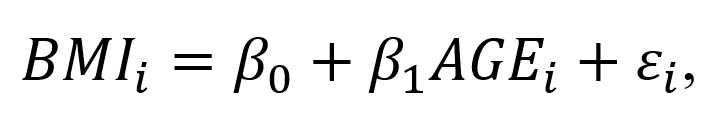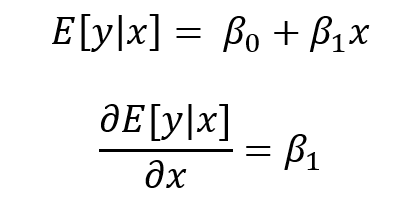In Stata, users have a lot of flexibility with creating plots, particularly after the margins command has been executed. Once a regression command has been run, users can estimate the average marginal effect of a factor with respect to another variable using the margins command in Stata. Once the average marginal effect has been estimated, users can plot this using the marginsplot or mplotoffset commands. These are power tools that allow us to visualize the average marginal effects, particularly when we have interaction terms.
I posted a tutorail on my RPubs site that revieweed some basic features of the marginsplot and mplotoffset commands and provide some practical examples of customization.